Run a set of models to generate fishery yield curve data for either planktivorous or demersal fish.
e2ep_run_ycurve.RdPerform a set of StrathE2E model runs along a sequence of values of either planktivorous or demersal fish harvest ratio multiplier, saving the annual average whole-domain biomass, annual landings and annual discards of all exploitable food web guilds from each run plus the annual average biomasses of all the other living components of the food web.
Usage
e2ep_run_ycurve(
model,
selection,
nyears = 50,
HRvector = c(0, 0.5, 1, 1.5, 2, 2.5, 3),
HRfixed = 1,
csv.output = FALSE
)Arguments
- model
R-list object defining a baseline model and configuration compiled by the e2ep_read() function.
- selection
Text string from a list identifying the fish guild for which a yield curve is to be generated. Select from: "PLANKTIV", "DEMERSAL", Remember to include the phrase within "" quotes.
- nyears
Number of years to run the StrathE2E model at each level of harvest ratio. Default = 50.
- HRvector
A vector of ascending order unique multiplier values to be applied to the baseline fish harvest ratio of the guild to be analysed. The values do not have to be evenly spaced (default = c(0,0.5,1.0,1.5,2.0,2.5,3.0)).
- HRfixed
Single value of the multiplier to be applied as the alternative (planktivorous or demersal) fish harvest ratio for all runs (default=1.0).
- csv.output
Logical. If TRUE then enable writing of csv output files (default=FALSE).
Value
Dataframe of annual average biomss, annual landings and annual discards of all exploitable guilds, plus average biomasses of other food web components, for each level of harvest ratio multiplier.
Details
The baseline for the sequence of runs (harvest ratio multiplier = 1.0) is a model name and variant as loaded by the e2ep_read() function.
The planktivorous or demersal fish yield curve can be generated for a given fixed setting of the other (demersal or planktivorous) fish harvest ratio multiplier (default = 1.0). All other conditions are held constant as in the baseline model configuration.
The yield curve represents the catch that would be generated from the stationary state of the model attained with long-term repeating annual cycles of all driving data. Hence it is important that each simulation is run for long enough that the model attains its stationary state, which may be some distance from the baseline model initial conditions. It is recommended that each run is at least 50 years.
The data on annual average biomass and annual integrated catches stored in the returned data object (and optionally a csv file) can subsequently be plotted using the function e2ep_plot_ycurve(). Users can easily plot any of the other saved data using their own plotting code.
If csv output is selected then the resulting files in the current user results folder have names Yield_curve_data_PFHRmult-*.csv or Yield_curve_data_DFHRmult-*.csv, depending on the 'selection' argument, and * represents the model.ident text set in the prior e2ep_read() function call.
Examples
# \donttest{
# Load the 2011-2019 version of the Barents Sea model supplied with the package :
model <- e2ep_read("Barents_Sea", "2011-2019", model.ident="11-19base")
#> Current working directory is...
#> 'C:/Users/jackl/OneDrive - University of Strathclyde/Documents/StrathE2E/strath-e-2-e-polar-webdev/docs/reference'
#> No 'results.path' specified so any csv data requested
#> will be directed to/from the temporary directory...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpSgdWsc'
#>
#> Model setup and parameters gathered from ...
#> StrathE2E2 package folder
#> Model results will be directed to/from ...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpSgdWsc/Barents_Sea/2011-2019/'
# In this example csv output is directed to a temporary folder since results.path is not set.
# In this illustrative example the model is run for only 3 years to enable quick
# return of results. In a real simulation nyear would be at least 50.
# This example illustrates that the vector of planktivorous fish harvest ratio multiplers
# does not have to be evenly spaced.
hr=c(0,0.5,0.75,1.0,1.25,2.0,3.0)
pf_yield_data <- e2ep_run_ycurve(model,selection="PLANKTIV", nyears=3, HRvector=hr,
HRfixed=1,csv.output=FALSE)
#> Run 1; pf harvest ratio multiplier = 0
#> Run 2; pf harvest ratio multiplier = 0.5
#> Run 3; pf harvest ratio multiplier = 0.75
#> Run 4; pf harvest ratio multiplier = 1
#> Run 5; pf harvest ratio multiplier = 1.25
#> Run 6; pf harvest ratio multiplier = 2
#> Run 7; pf harvest ratio multiplier = 3
# View the column names of the results dataframe:
names(pf_yield_data)
#> [1] "PlankFishHRmult" "DemFishHRmult" "PlankFishbiom"
#> [4] "PlankFishland" "PlankFishdisc" "DemFishbiom"
#> [7] "DemFishland" "DemFishdisc" "MigFishbiom"
#> [10] "MigFishland" "MigFishdisc" "Benthsuspdepbiom"
#> [13] "Benthsuspdepland" "Benthsuspdepdisc" "Benthcarnscavbiom"
#> [16] "Benthcarnscavland" "Benthcarnscavdisc" "CarnivZoobiom"
#> [19] "CarnivZooland" "CarnivZoodisc" "Birdbiom"
#> [22] "Birdland" "Birddisc" "Pinnipedbiom"
#> [25] "Pinnipedland" "Pinnipeddisc" "Cetaceanbiom"
#> [28] "Cetaceanland" "Cetaceandisc" "Macrophytebiom"
#> [31] "Macrophyteland" "Macrophytedisc" "Icealgaebiom"
#> [34] "Phytoplanktonbiom" "OmnivZoobiom" "MaritimeMambiom"
# Plotting the results...
# The planktivorous fish yield curve can be plotted using the function:
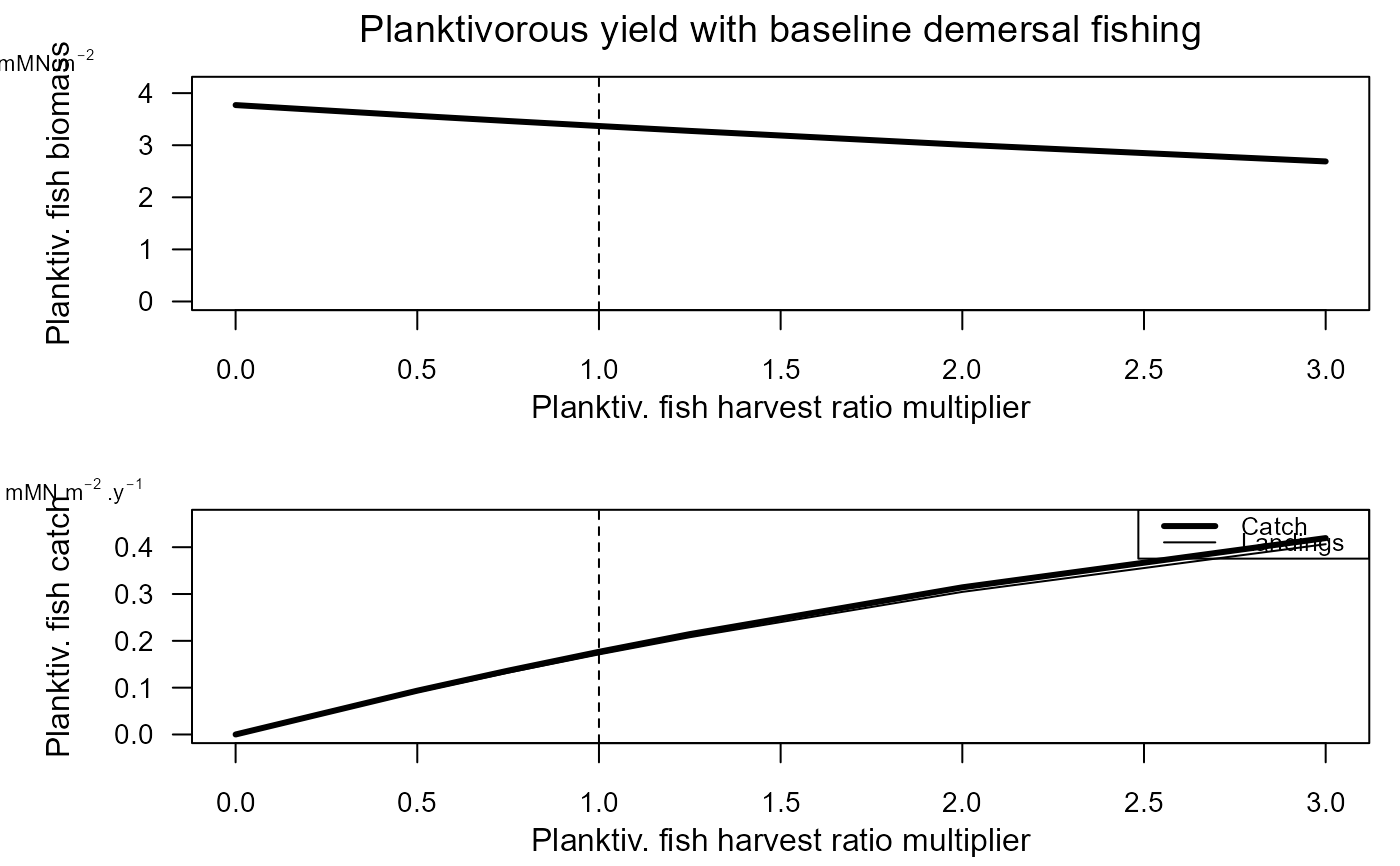
e2ep_plot_ycurve(model, selection="PLANKTIV", results=pf_yield_data,
title="Planktivorous yield with baseline demersal fishing")
 #> PlankFishHRmult DemFishHRmult PlankFishbiom PlankFishland PlankFishdisc
#> 1 0.00 1 3.770675 0.00000000 0.000000000
#> 2 0.50 1 3.564086 0.09064502 0.002905956
#> 3 0.75 1 3.465040 0.13207980 0.004234280
#> 4 1.00 1 3.368731 0.17106605 0.005484099
#> 5 1.25 1 3.275091 0.20770665 0.006658714
#> 6 2.00 1 3.009514 0.30453202 0.009762703
#> 7 3.00 1 2.688818 0.40644554 0.013029850
#> DemFishbiom DemFishland DemFishdisc MigFishbiom MigFishland MigFishdisc
#> 1 7.293379 0.3161697 0.01026231 0.06320250 0.001604206 5.854998e-07
#> 2 7.295125 0.3162420 0.01026432 0.06320490 0.001604164 5.854845e-07
#> 3 7.295981 0.3162774 0.01026531 0.06320434 0.001604197 5.854964e-07
#> 4 7.296815 0.3163120 0.01026627 0.06320504 0.001604326 5.855436e-07
#> 5 7.297623 0.3163454 0.01026720 0.06320356 0.001604238 5.855114e-07
#> 6 7.299966 0.3164425 0.01026991 0.06320565 0.001604315 5.855394e-07
#> 7 7.302888 0.3165637 0.01027329 0.06320775 0.001604367 5.855585e-07
#> Benthsuspdepbiom Benthsuspdepland Benthsuspdepdisc Benthcarnscavbiom
#> 1 51.46423 2.206148e-07 0 8.755223
#> 2 51.44646 2.204815e-07 0 8.753275
#> 3 51.43789 2.204170e-07 0 8.752334
#> 4 51.42955 2.203543e-07 0 8.751418
#> 5 51.42141 2.202934e-07 0 8.750523
#> 6 51.39814 2.201186e-07 0 8.747960
#> 7 51.36970 2.199053e-07 0 8.744813
#> Benthcarnscavland Benthcarnscavdisc CarnivZoobiom CarnivZooland CarnivZoodisc
#> 1 0.01525942 0 9.293020 1.561344e-07 0
#> 2 0.01525540 0 9.306990 1.565387e-07 0
#> 3 0.01525345 0 9.313733 1.567339e-07 0
#> 4 0.01525156 0 9.320318 1.569245e-07 0
#> 5 0.01524971 0 9.326749 1.571106e-07 0
#> 6 0.01524442 0 9.345156 1.576434e-07 0
#> 7 0.01523792 0 9.367740 1.582971e-07 0
#> Birdbiom Birdland Birddisc Pinnipedbiom Pinnipedland Pinnipeddisc
#> 1 0.01254918 0 1.452183e-05 0.07208542 0.0001355529 1.009320e-05
#> 2 0.01245810 0 1.441165e-05 0.07178199 0.0001376286 1.005783e-05
#> 3 0.01241409 0 1.435843e-05 0.07163500 0.0001386695 1.004080e-05
#> 4 0.01237112 0 1.430648e-05 0.07149120 0.0001397117 1.002422e-05
#> 5 0.01232905 0 1.425562e-05 0.07135038 0.0001407546 1.000806e-05
#> 6 0.01220866 0 1.411005e-05 0.07094560 0.0001438823 9.961996e-06
#> 7 0.01206049 0 1.393091e-05 0.07044475 0.0001480303 9.905870e-06
#> Cetaceanbiom Cetaceanland Cetaceandisc Macrophytebiom Macrophyteland
#> 1 0.4539863 0.0009073163 2.390269e-05 50.41281 0
#> 2 0.4536256 0.0009065412 2.388267e-05 50.41329 0
#> 3 0.4534506 0.0009061652 2.387296e-05 50.41353 0
#> 4 0.4532801 0.0009057981 2.386348e-05 50.41375 0
#> 5 0.4531093 0.0009054319 2.385404e-05 50.41398 0
#> 6 0.4526244 0.0009043881 2.382713e-05 50.41462 0
#> 7 0.4520205 0.0009030864 2.379361e-05 50.41541 0
#> Macrophytedisc Icealgaebiom Phytoplanktonbiom OmnivZoobiom MaritimeMambiom
#> 1 0 0.06400509 8.303649 94.59465 0.002518150
#> 2 0 0.06400147 8.302425 94.65797 0.002517576
#> 3 0 0.06399994 8.301837 94.68843 0.002517302
#> 4 0 0.06399812 8.301266 94.71809 0.002517006
#> 5 0 0.06399646 8.300710 94.74696 0.002516730
#> 6 0 0.06399197 8.299129 94.82921 0.002515903
#> 7 0 0.06398618 8.297214 94.92916 0.002514871
# }
#> PlankFishHRmult DemFishHRmult PlankFishbiom PlankFishland PlankFishdisc
#> 1 0.00 1 3.770675 0.00000000 0.000000000
#> 2 0.50 1 3.564086 0.09064502 0.002905956
#> 3 0.75 1 3.465040 0.13207980 0.004234280
#> 4 1.00 1 3.368731 0.17106605 0.005484099
#> 5 1.25 1 3.275091 0.20770665 0.006658714
#> 6 2.00 1 3.009514 0.30453202 0.009762703
#> 7 3.00 1 2.688818 0.40644554 0.013029850
#> DemFishbiom DemFishland DemFishdisc MigFishbiom MigFishland MigFishdisc
#> 1 7.293379 0.3161697 0.01026231 0.06320250 0.001604206 5.854998e-07
#> 2 7.295125 0.3162420 0.01026432 0.06320490 0.001604164 5.854845e-07
#> 3 7.295981 0.3162774 0.01026531 0.06320434 0.001604197 5.854964e-07
#> 4 7.296815 0.3163120 0.01026627 0.06320504 0.001604326 5.855436e-07
#> 5 7.297623 0.3163454 0.01026720 0.06320356 0.001604238 5.855114e-07
#> 6 7.299966 0.3164425 0.01026991 0.06320565 0.001604315 5.855394e-07
#> 7 7.302888 0.3165637 0.01027329 0.06320775 0.001604367 5.855585e-07
#> Benthsuspdepbiom Benthsuspdepland Benthsuspdepdisc Benthcarnscavbiom
#> 1 51.46423 2.206148e-07 0 8.755223
#> 2 51.44646 2.204815e-07 0 8.753275
#> 3 51.43789 2.204170e-07 0 8.752334
#> 4 51.42955 2.203543e-07 0 8.751418
#> 5 51.42141 2.202934e-07 0 8.750523
#> 6 51.39814 2.201186e-07 0 8.747960
#> 7 51.36970 2.199053e-07 0 8.744813
#> Benthcarnscavland Benthcarnscavdisc CarnivZoobiom CarnivZooland CarnivZoodisc
#> 1 0.01525942 0 9.293020 1.561344e-07 0
#> 2 0.01525540 0 9.306990 1.565387e-07 0
#> 3 0.01525345 0 9.313733 1.567339e-07 0
#> 4 0.01525156 0 9.320318 1.569245e-07 0
#> 5 0.01524971 0 9.326749 1.571106e-07 0
#> 6 0.01524442 0 9.345156 1.576434e-07 0
#> 7 0.01523792 0 9.367740 1.582971e-07 0
#> Birdbiom Birdland Birddisc Pinnipedbiom Pinnipedland Pinnipeddisc
#> 1 0.01254918 0 1.452183e-05 0.07208542 0.0001355529 1.009320e-05
#> 2 0.01245810 0 1.441165e-05 0.07178199 0.0001376286 1.005783e-05
#> 3 0.01241409 0 1.435843e-05 0.07163500 0.0001386695 1.004080e-05
#> 4 0.01237112 0 1.430648e-05 0.07149120 0.0001397117 1.002422e-05
#> 5 0.01232905 0 1.425562e-05 0.07135038 0.0001407546 1.000806e-05
#> 6 0.01220866 0 1.411005e-05 0.07094560 0.0001438823 9.961996e-06
#> 7 0.01206049 0 1.393091e-05 0.07044475 0.0001480303 9.905870e-06
#> Cetaceanbiom Cetaceanland Cetaceandisc Macrophytebiom Macrophyteland
#> 1 0.4539863 0.0009073163 2.390269e-05 50.41281 0
#> 2 0.4536256 0.0009065412 2.388267e-05 50.41329 0
#> 3 0.4534506 0.0009061652 2.387296e-05 50.41353 0
#> 4 0.4532801 0.0009057981 2.386348e-05 50.41375 0
#> 5 0.4531093 0.0009054319 2.385404e-05 50.41398 0
#> 6 0.4526244 0.0009043881 2.382713e-05 50.41462 0
#> 7 0.4520205 0.0009030864 2.379361e-05 50.41541 0
#> Macrophytedisc Icealgaebiom Phytoplanktonbiom OmnivZoobiom MaritimeMambiom
#> 1 0 0.06400509 8.303649 94.59465 0.002518150
#> 2 0 0.06400147 8.302425 94.65797 0.002517576
#> 3 0 0.06399994 8.301837 94.68843 0.002517302
#> 4 0 0.06399812 8.301266 94.71809 0.002517006
#> 5 0 0.06399646 8.300710 94.74696 0.002516730
#> 6 0 0.06399197 8.299129 94.82921 0.002515903
#> 7 0 0.06398618 8.297214 94.92916 0.002514871
# }