Create tornado bar-plots of the differences between two model runs.
e2e_compare_runs_bar.RdCreate a tornado bar-plot diagram of the differences in either annual average masses of ecology model varibles, or annual fishery catches, between two different runs of the StrathE2E model, referred to as baseline and sceanrio runs.
Usage
e2e_compare_runs_bar(
selection = "AAM",
model1 = NA,
use.saved1 = FALSE,
results1,
model2 = NA,
use.saved2 = FALSE,
results2,
log.pc = "PC",
zone = "W",
bpmin = (-50),
bpmax = (+50),
maintitle = ""
)Arguments
- selection
Text string from a list identifying which type of data are to be plotted. Select from: "AAM", "CATCH", corresponding to annual average mass data, and catch data respectively (default = "AAM"). Remember to include the phrase within "" quotes.
- model1
R-list object defining the baseline model configuration compiled by the e2e_read() function.
- use.saved1
Logical. If TRUE then use baseline data from a prior user-defined run held as csv files data in the current results folder (default=FALSE).
- results1
R-list object of baseline model output generated by the e2e_run() (default=NULL).
- model2
R-list object defining the scenario model configuration compiled by the e2e_read() function.
- use.saved2
Logical. If TRUE then use scenario data from a prior user-defined run held as csv files data in the current results folder (default=FALSE).
- results2
R-list object of scenario model output generated by the e2e_run() (default=NULL).
- log.pc
Value="LG" for data to be plotted on a log10 scale, value = "PC" for data to be plotted on a percentage difference scale (default = "PC").
- zone
Value = "O" for offshore, "I" for inshore, or "W" for whole model domain (all upper case) (default = "W").
- bpmin
Axis minimum for plot - i.e. the maximum NEGATIVE value of (scenario-baseline). Default = -50, given log.pc="PC" (percentage differences). Needs to be reset to e.g. -0.3 if log.pc="LG" (log scale).
- bpmax
Axis maximum for plot - i.e. the maximum POSITIVE value of (scenario-baseline). Default = +50, given log.pc="PC" (percentage differences). Needs to be reset to e.g. +0.3 if log.pc="LG" (log scale).
- maintitle
A optional descriptive text field (in quotes) to be added above the plot. Keep to 45 characters including spaces (default="").
Value
List object comprising two dataframes of the displayed data, graphical display in a new graphics window.
Details
A tornado plot is a horizontal barplot which shows the difference between baseline and sceanrio runs. Bars to the right indicate scenario values greater than the baseline, bars to the left indicate scenario values less than baseline. In this function the difference between scenario and baseline data is calculated as either ((scenario - baseline)/baseline) and plotted on a linear (percentage) scale, or (scenario/baseline) and plotted on a log10 scale. In both cases, zero represents scenario = baseline.
The choice of whether to plot data on the masses of ecosystem components, or fishery catches, is determined by an argument setting in the function call. In either case two panels of tornado diagrams are plotted. In the case of mass data, the panels show data on water column components of the ecoststem (upper panel) and seabed components of the system (lower panel). In the case of catch data, the upper panel shows landings, the lower shows discards. For mass data and landings, positive values (scenario > baseline) are indicated by green bars to the right, negative values (scenario < baseline) by red bars to the left. For discards, positive values are in black, negative in grey. For any bars extending outsided the selected plotting range, the numeric value is displayed inside the relevant bar.
The function accepts data from either saved csv files created by the e2e_run() function, or directly from e2e_run() results objects. The relevant csv files are: /results/Modelname/Variantname/ZONE_model_anav_biomass-*.csv where ZONE is INSHORE, OFFSHORE or WHOLEDOMAIN and * represents the model run identifier (model.ident) embedded in the R-list object created by the e2e_read() function.
In addition to generating graphics output the function returns a list object of the data presented in the plot. The object comprises two dataframes (changewater and changeseabed where annual average mass data are selected; changeland and changedisc where catch data are selected). The first column in each dataframe is the proportional difference expressed on a log10 scale, the second column as a percentage.
Examples
# Load the 1970-1999 version of the internal North Sea model and run for 1 year
m1 <- e2e_read("North_Sea", "1970-1999",model.ident="70_99")
#> Current working directory is...
#> 'C:/Users/jackl/OneDrive - University of Strathclyde/Documents/Research/Packages/StrathE2E/strathe2e2/docs/reference'
#> No 'results.path' specified so any csv data requested
#> will be directed to/from the temporary directory...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpYdRhoX'
#>
#> Model setup and parameters gathered from ...
#> StrathE2E2 package folder
#> Model results will be directed to/from ...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpYdRhoX/North_Sea/1970-1999/'
r1 <-e2e_run(m1,nyears=1)
# Load the 2003-2013 version of the internal North Sea model and run for 1 year
m2 <- e2e_read("North_Sea", "2003-2013",model.ident="03_13")
#> Current working directory is...
#> 'C:/Users/jackl/OneDrive - University of Strathclyde/Documents/Research/Packages/StrathE2E/strathe2e2/docs/reference'
#> No 'results.path' specified so any csv data requested
#> will be directed to/from the temporary directory...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpYdRhoX'
#>
#> Model setup and parameters gathered from ...
#> StrathE2E2 package folder
#> Model results will be directed to/from ...
#> 'C:\Users\jackl\AppData\Local\Temp\RtmpYdRhoX/North_Sea/2003-2013/'
r2 <-e2e_run(m2,nyears=1)
# Compare the annual average mass in 1970-1999 (as the baseline case) with 2003-2013
# (as the scenario case):
mdiff_results1 <- e2e_compare_runs_bar(selection="AAM",
model1=NA, use.saved1=FALSE, results1=r1,
model2=NA, use.saved2=FALSE, results2=r2,
log.pc="PC", zone="W",
bpmin=(-50),bpmax=(+50),
maintitle="2003-2013 compared with 1970-1999")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in memory from an existing model run"
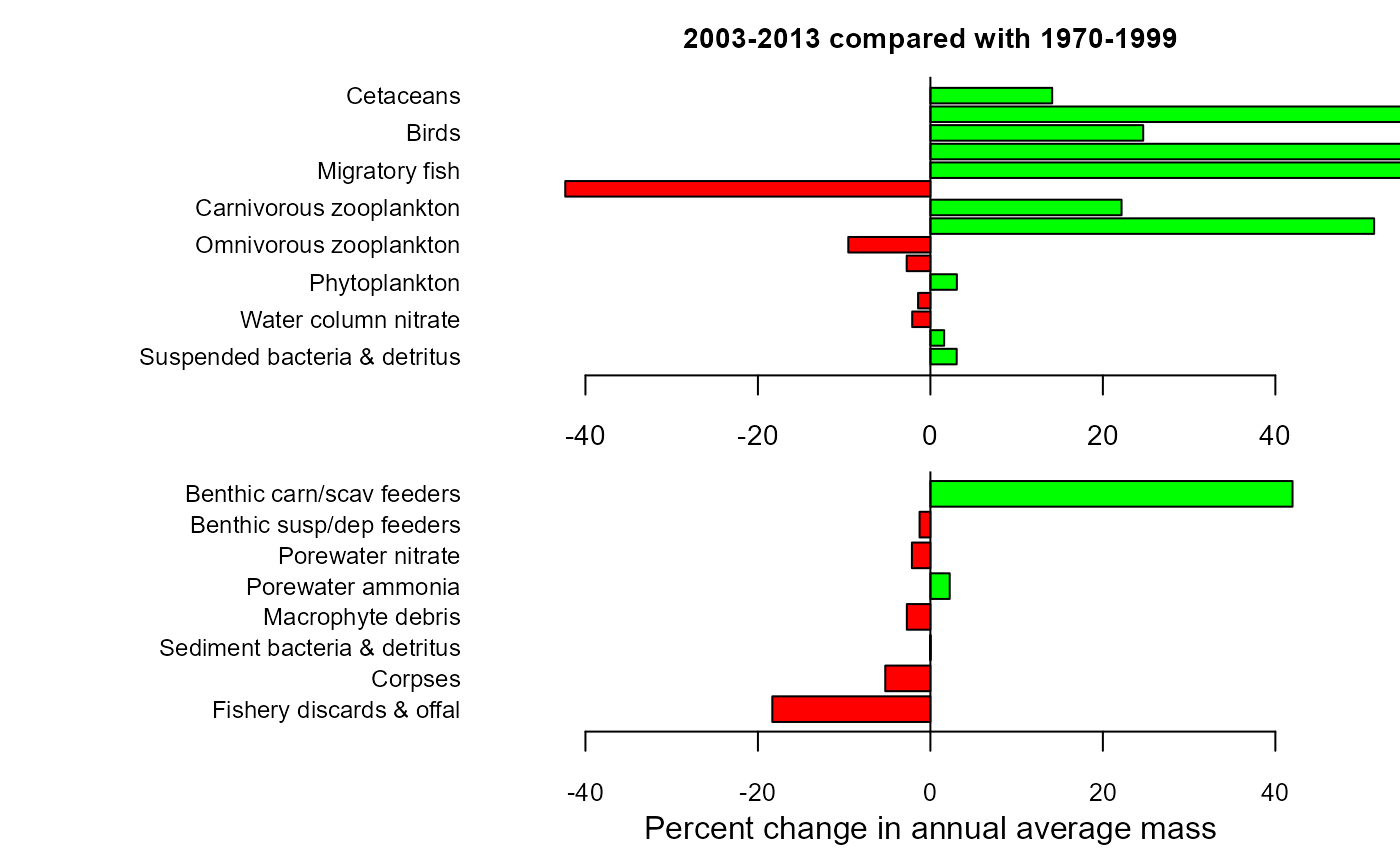 mdiff_results1
#> $changewater
#> LG PC
#> Suspended bacteria & detritus 0.013044733 3.049226
#> Water column ammonia 0.006868140 1.594019
#> Water column nitrate -0.009261893 -2.110050
#> Macrophytes -0.006248559 -1.428483
#> Phytoplankton 0.013128237 3.069041
#> Benthic susp/dep feeder larvae -0.012110314 -2.749983
#> Omnivorous zooplankton -0.043449013 -9.520334
#> Benthic carn/scav feeder larvae 0.180277050 51.452711
#> Carnivorous zooplankton 0.086949334 22.165713
#> Plantiv. fish adults+larvae -0.239130983 -42.340746
#> Migratory fish 0.409867893 156.961402
#> Demersal fish adults+larvae 0.463824135 190.953868
#> Birds 0.095787717 24.677394
#> Pinnipeds 0.543189753 249.292896
#> Cetaceans 0.057366103 14.121140
#>
#> $changeseabed
#> LG PC
#> Fishery discards & offal -8.794381e-02 -18.331197378
#> Corpses -2.334533e-02 -5.233538354
#> Sediment bacteria & detritus 1.111604e-05 0.002559595
#> Macrophyte debris -1.202910e-02 -2.731795999
#> Porewater ammonia 9.622150e-03 2.240308192
#> Porewater nitrate -9.414269e-03 -2.144389464
#> Benthic susp/dep feeders -5.489552e-03 -1.256060835
#> Benthic carn/scav feeders 1.522613e-01 41.991163821
#>
# Compare the annual catch in 1970-1999 (as the baseline case) with 2003-2013
# (as the scenario case):
mdiff_results2 <- e2e_compare_runs_bar(selection="CATCH",
model1=NA, use.saved1=FALSE, results1=r1,
model2=NA, use.saved2=FALSE, results2=r2,
log.pc="LG", zone="W",
bpmin=(-0.9),bpmax=(+0.4),
maintitle="2003-2013 compared with 1970-1999")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in memory from an existing model run"
mdiff_results1
#> $changewater
#> LG PC
#> Suspended bacteria & detritus 0.013044733 3.049226
#> Water column ammonia 0.006868140 1.594019
#> Water column nitrate -0.009261893 -2.110050
#> Macrophytes -0.006248559 -1.428483
#> Phytoplankton 0.013128237 3.069041
#> Benthic susp/dep feeder larvae -0.012110314 -2.749983
#> Omnivorous zooplankton -0.043449013 -9.520334
#> Benthic carn/scav feeder larvae 0.180277050 51.452711
#> Carnivorous zooplankton 0.086949334 22.165713
#> Plantiv. fish adults+larvae -0.239130983 -42.340746
#> Migratory fish 0.409867893 156.961402
#> Demersal fish adults+larvae 0.463824135 190.953868
#> Birds 0.095787717 24.677394
#> Pinnipeds 0.543189753 249.292896
#> Cetaceans 0.057366103 14.121140
#>
#> $changeseabed
#> LG PC
#> Fishery discards & offal -8.794381e-02 -18.331197378
#> Corpses -2.334533e-02 -5.233538354
#> Sediment bacteria & detritus 1.111604e-05 0.002559595
#> Macrophyte debris -1.202910e-02 -2.731795999
#> Porewater ammonia 9.622150e-03 2.240308192
#> Porewater nitrate -9.414269e-03 -2.144389464
#> Benthic susp/dep feeders -5.489552e-03 -1.256060835
#> Benthic carn/scav feeders 1.522613e-01 41.991163821
#>
# Compare the annual catch in 1970-1999 (as the baseline case) with 2003-2013
# (as the scenario case):
mdiff_results2 <- e2e_compare_runs_bar(selection="CATCH",
model1=NA, use.saved1=FALSE, results1=r1,
model2=NA, use.saved2=FALSE, results2=r2,
log.pc="LG", zone="W",
bpmin=(-0.9),bpmax=(+0.4),
maintitle="2003-2013 compared with 1970-1999")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in memory from an existing model run"
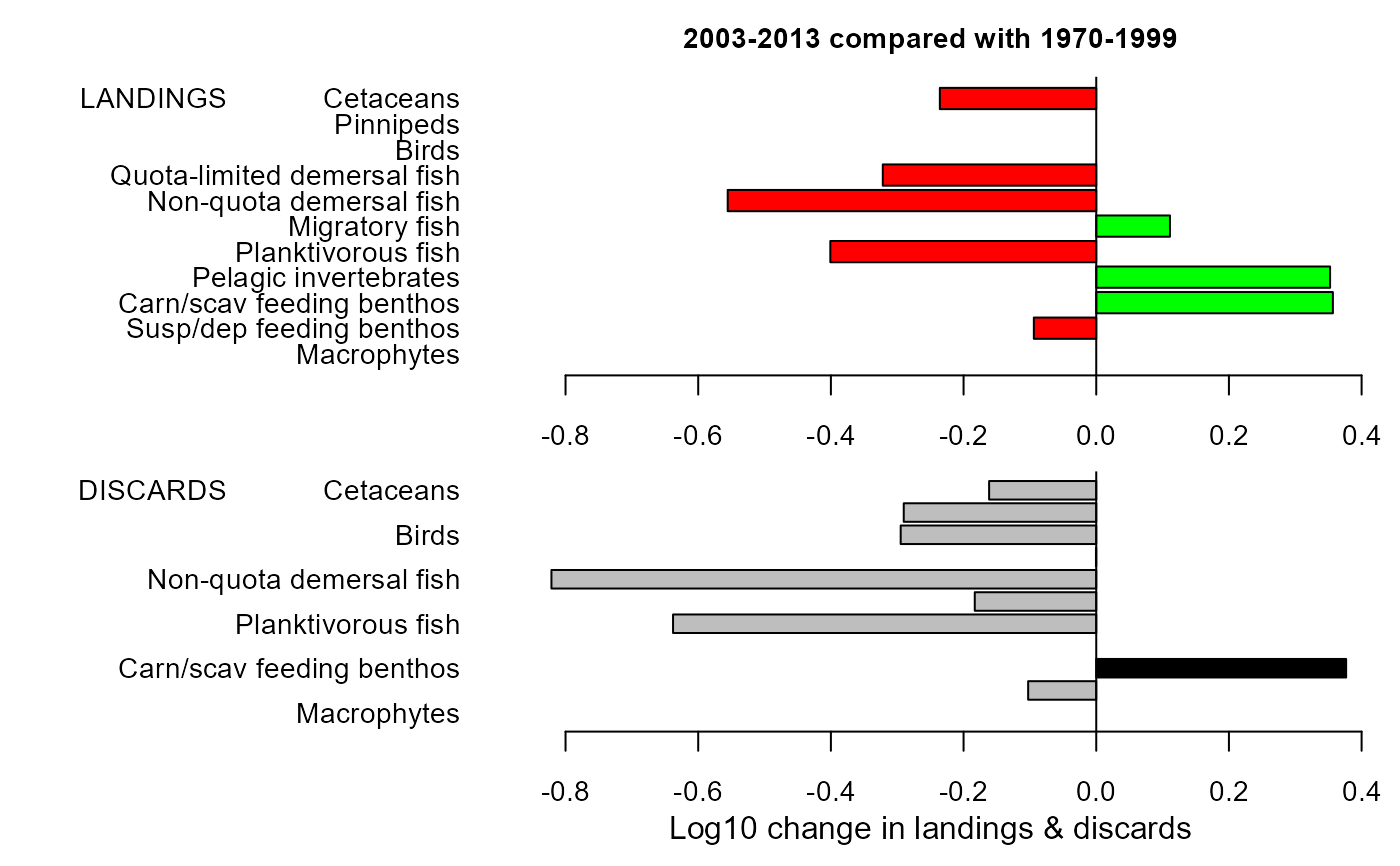 mdiff_results2
#> $changeland
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.09406901 -19.47495
#> Carn/scav feeding benthos 0.35670443 127.35496
#> Pelagic invertebrates 0.35249546 125.16219
#> Planktivorous fish -0.40085508 -60.26759
#> Migratory fish 0.11111679 29.15666
#> Non-quota demersal fish -0.55555150 -72.17415
#> Quota-limited demersal fish -0.32177121 -52.33180
#> Birds NA NA
#> Pinnipeds NA NA
#> Cetaceans -0.23575081 -41.89023
#>
#> $changedisc
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.1026395412 -21.0484867
#> Carn/scav feeding benthos 0.3766875813 138.0606315
#> Pelagic invertebrates NA NA
#> Planktivorous fish -0.6379264830 -76.9816856
#> Migratory fish -0.1831228843 -34.4040365
#> Non-quota demersal fish -0.8212578829 -84.9081626
#> Quota-limited demersal fish -0.0004238928 -0.0975573
#> Birds -0.2947809986 -49.2753567
#> Pinnipeds -0.2901815803 -48.7353001
#> Cetaceans -0.1614880120 -31.0535376
#>
# \donttest{
# Create a new scenario run from 2003-2013 case.
# Copy the 2003-2013 configuration into a new model object:
scen1_model <- m2
scen1_model$setup$model.ident <- "scenario1"
# Gear 4 (Beam_Trawl_BT1+BT2) activity rate rescaled to 0.5*baseline:
scen1_model$data$fleet.model$gear_mult[4] <- 0.5
scen1_results <- e2e_run(scen1_model,nyears=20)
# Compare the annual average mass from the the 2003-2013 baseline with scenario1 data
mdiff_results3 <- e2e_compare_runs_bar(selection="AAM",
model1=NA, use.saved1=FALSE, results1=r2,
model2=NA, use.saved2=FALSE, results2=scen1_results,
log.pc="PC", zone="W",
bpmin=(-30),bpmax=(+30),
maintitle="Beam Trawl activity reduced by half")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in memory from an existing model run"
mdiff_results2
#> $changeland
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.09406901 -19.47495
#> Carn/scav feeding benthos 0.35670443 127.35496
#> Pelagic invertebrates 0.35249546 125.16219
#> Planktivorous fish -0.40085508 -60.26759
#> Migratory fish 0.11111679 29.15666
#> Non-quota demersal fish -0.55555150 -72.17415
#> Quota-limited demersal fish -0.32177121 -52.33180
#> Birds NA NA
#> Pinnipeds NA NA
#> Cetaceans -0.23575081 -41.89023
#>
#> $changedisc
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.1026395412 -21.0484867
#> Carn/scav feeding benthos 0.3766875813 138.0606315
#> Pelagic invertebrates NA NA
#> Planktivorous fish -0.6379264830 -76.9816856
#> Migratory fish -0.1831228843 -34.4040365
#> Non-quota demersal fish -0.8212578829 -84.9081626
#> Quota-limited demersal fish -0.0004238928 -0.0975573
#> Birds -0.2947809986 -49.2753567
#> Pinnipeds -0.2901815803 -48.7353001
#> Cetaceans -0.1614880120 -31.0535376
#>
# \donttest{
# Create a new scenario run from 2003-2013 case.
# Copy the 2003-2013 configuration into a new model object:
scen1_model <- m2
scen1_model$setup$model.ident <- "scenario1"
# Gear 4 (Beam_Trawl_BT1+BT2) activity rate rescaled to 0.5*baseline:
scen1_model$data$fleet.model$gear_mult[4] <- 0.5
scen1_results <- e2e_run(scen1_model,nyears=20)
# Compare the annual average mass from the the 2003-2013 baseline with scenario1 data
mdiff_results3 <- e2e_compare_runs_bar(selection="AAM",
model1=NA, use.saved1=FALSE, results1=r2,
model2=NA, use.saved2=FALSE, results2=scen1_results,
log.pc="PC", zone="W",
bpmin=(-30),bpmax=(+30),
maintitle="Beam Trawl activity reduced by half")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in memory from an existing model run"
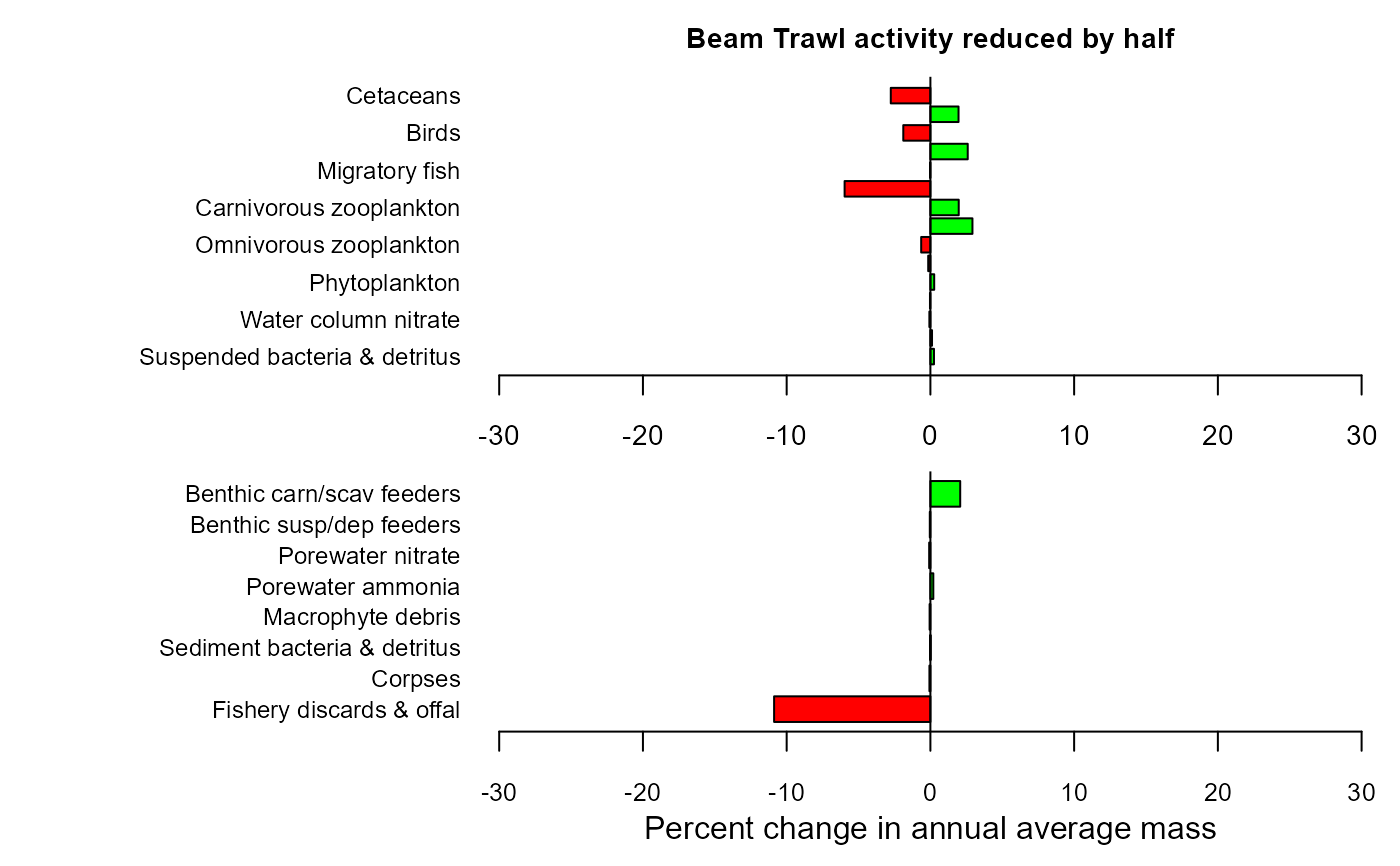 mdiff_results3
#> $changewater
#> LG PC
#> Suspended bacteria & detritus 0.0010795507 0.24888494
#> Water column ammonia 0.0004891751 0.11270019
#> Water column nitrate -0.0003095694 -0.07125558
#> Macrophytes -0.0001301325 -0.02995963
#> Phytoplankton 0.0011463468 0.26430479
#> Benthic susp/dep feeder larvae -0.0006611424 -0.15211784
#> Omnivorous zooplankton -0.0027936496 -0.64119709
#> Benthic carn/scav feeder larvae 0.0125216916 2.92519336
#> Carnivorous zooplankton 0.0084754045 1.97070092
#> Plantiv. fish adults+larvae -0.0267297274 -5.96916934
#> Migratory fish -0.0001061136 -0.02443057
#> Demersal fish adults+larvae 0.0111341117 2.59687002
#> Birds -0.0082693152 -1.88606708
#> Pinnipeds 0.0084173714 1.95707586
#> Cetaceans -0.0121374528 -2.75605988
#>
#> $changeseabed
#> LG PC
#> Fishery discards & offal -4.999510e-02 -1.087390e+01
#> Corpses -2.941530e-04 -6.770830e-02
#> Sediment bacteria & detritus 3.572280e-06 8.225512e-04
#> Macrophyte debris -2.382308e-04 -5.483963e-02
#> Porewater ammonia 8.792748e-04 2.026656e-01
#> Porewater nitrate -3.695883e-04 -8.506466e-02
#> Benthic susp/dep feeders -1.817597e-04 -4.184297e-02
#> Benthic carn/scav feeders 8.913719e-03 2.073667e+00
#>
# }
# \donttest{
# Create a second sceanario from the 1970-1999 case, this time saving the results to a file;
# csv output to temporary folder since results.path was not set in e2e_read() when creating m1.
# Copy the baseline configuration into a new model object:
scen2_model <- m1
scen2_model$setup$model.ident <- "scenario2"
# Gear 1 (Pelagic_Trawl+Seine) activity rate rescaled to 0.5*baseline:
scen2_model$data$fleet.model$gear_mult[1] <- 0.5
scen2_results <- e2e_run(scen2_model,nyears=20, csv.output=TRUE)
# Compare the annual catches in the 1970-1999 base line with the Pelagic Trawl/seine scenario
mdiff_results4 <- e2e_compare_runs_bar(selection="CATCH",
model1=NA, use.saved1=FALSE, results1=r1,
model2=scen2_model,use.saved2=TRUE, results2=NA,
log.pc="LG", zone="W",
bpmin=(-0.4),bpmax=(+0.6),
maintitle="Pelagic Trawl/Seine activity reduced by half")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in a csv files from a past model run"
mdiff_results3
#> $changewater
#> LG PC
#> Suspended bacteria & detritus 0.0010795507 0.24888494
#> Water column ammonia 0.0004891751 0.11270019
#> Water column nitrate -0.0003095694 -0.07125558
#> Macrophytes -0.0001301325 -0.02995963
#> Phytoplankton 0.0011463468 0.26430479
#> Benthic susp/dep feeder larvae -0.0006611424 -0.15211784
#> Omnivorous zooplankton -0.0027936496 -0.64119709
#> Benthic carn/scav feeder larvae 0.0125216916 2.92519336
#> Carnivorous zooplankton 0.0084754045 1.97070092
#> Plantiv. fish adults+larvae -0.0267297274 -5.96916934
#> Migratory fish -0.0001061136 -0.02443057
#> Demersal fish adults+larvae 0.0111341117 2.59687002
#> Birds -0.0082693152 -1.88606708
#> Pinnipeds 0.0084173714 1.95707586
#> Cetaceans -0.0121374528 -2.75605988
#>
#> $changeseabed
#> LG PC
#> Fishery discards & offal -4.999510e-02 -1.087390e+01
#> Corpses -2.941530e-04 -6.770830e-02
#> Sediment bacteria & detritus 3.572280e-06 8.225512e-04
#> Macrophyte debris -2.382308e-04 -5.483963e-02
#> Porewater ammonia 8.792748e-04 2.026656e-01
#> Porewater nitrate -3.695883e-04 -8.506466e-02
#> Benthic susp/dep feeders -1.817597e-04 -4.184297e-02
#> Benthic carn/scav feeders 8.913719e-03 2.073667e+00
#>
# }
# \donttest{
# Create a second sceanario from the 1970-1999 case, this time saving the results to a file;
# csv output to temporary folder since results.path was not set in e2e_read() when creating m1.
# Copy the baseline configuration into a new model object:
scen2_model <- m1
scen2_model$setup$model.ident <- "scenario2"
# Gear 1 (Pelagic_Trawl+Seine) activity rate rescaled to 0.5*baseline:
scen2_model$data$fleet.model$gear_mult[1] <- 0.5
scen2_results <- e2e_run(scen2_model,nyears=20, csv.output=TRUE)
# Compare the annual catches in the 1970-1999 base line with the Pelagic Trawl/seine scenario
mdiff_results4 <- e2e_compare_runs_bar(selection="CATCH",
model1=NA, use.saved1=FALSE, results1=r1,
model2=scen2_model,use.saved2=TRUE, results2=NA,
log.pc="LG", zone="W",
bpmin=(-0.4),bpmax=(+0.6),
maintitle="Pelagic Trawl/Seine activity reduced by half")
#> [1] "Using baseline data held in memory from an existing model run"
#> [1] "Using scenario data held in a csv files from a past model run"
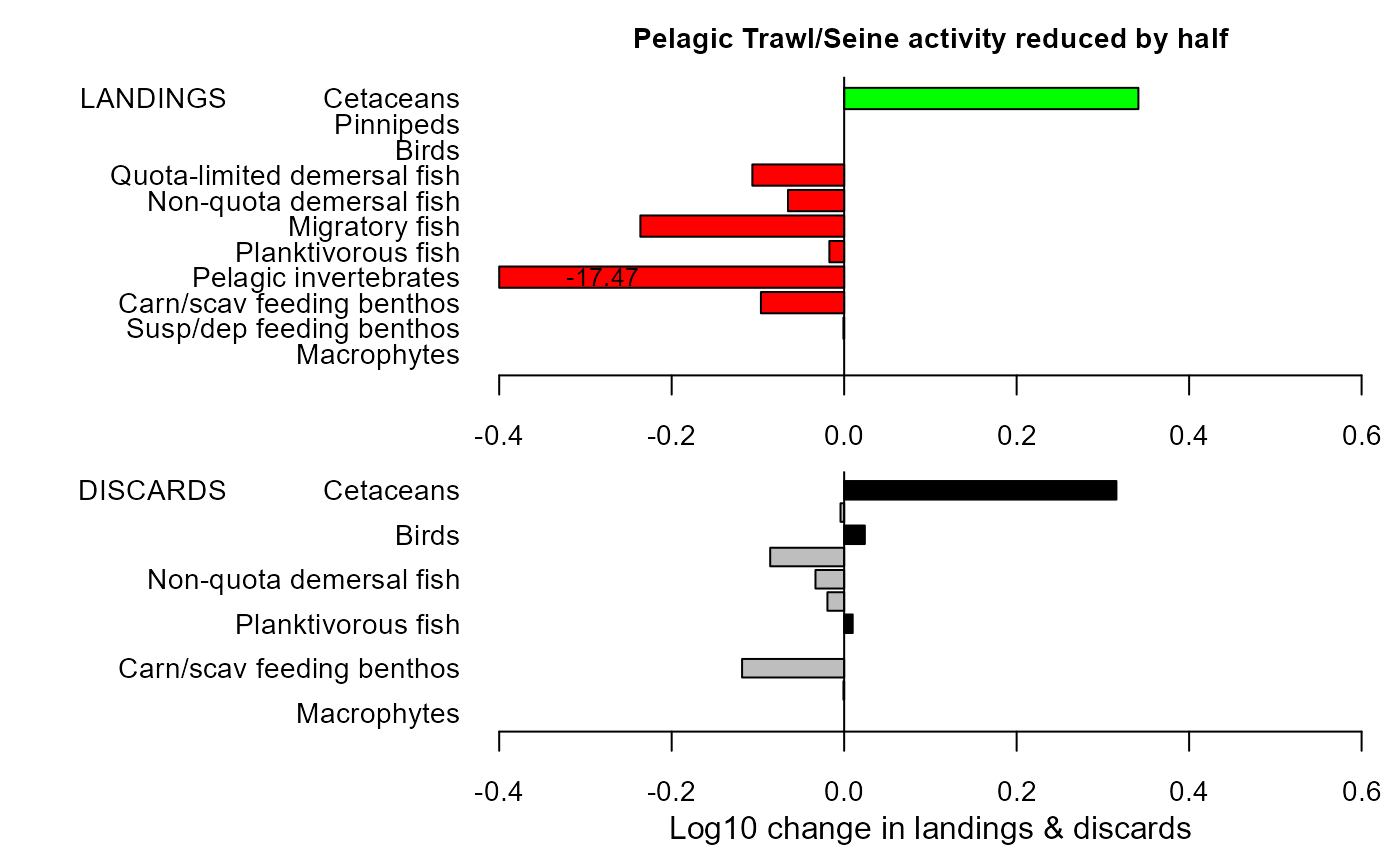 mdiff_results4
#> $changeland
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -8.477639e-04 -0.1950145
#> Carn/scav feeding benthos -9.652525e-02 -19.9290928
#> Pelagic invertebrates -1.746671e+01 -100.0000000
#> Planktivorous fish -1.715262e-02 -3.8725591
#> Migratory fish -2.363041e-01 -41.9642091
#> Non-quota demersal fish -6.512231e-02 -13.9248689
#> Quota-limited demersal fish -1.064561e-01 -21.7392630
#> Birds NA NA
#> Pinnipeds NA NA
#> Cetaceans 3.411624e-01 119.3624969
#>
#> $changedisc
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.0008184394 -0.1882752
#> Carn/scav feeding benthos -0.1183588979 -23.8550507
#> Pelagic invertebrates NA NA
#> Planktivorous fish 0.0098975832 2.3051704
#> Migratory fish -0.0193985593 -4.3683959
#> Non-quota demersal fish -0.0332555585 -7.3715405
#> Quota-limited demersal fish -0.0857658131 -17.9205974
#> Birds 0.0239826045 5.6775180
#> Pinnipeds -0.0042272205 -0.9686317
#> Cetaceans 0.3157041768 106.8731735
#>
# }
mdiff_results4
#> $changeland
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -8.477639e-04 -0.1950145
#> Carn/scav feeding benthos -9.652525e-02 -19.9290928
#> Pelagic invertebrates -1.746671e+01 -100.0000000
#> Planktivorous fish -1.715262e-02 -3.8725591
#> Migratory fish -2.363041e-01 -41.9642091
#> Non-quota demersal fish -6.512231e-02 -13.9248689
#> Quota-limited demersal fish -1.064561e-01 -21.7392630
#> Birds NA NA
#> Pinnipeds NA NA
#> Cetaceans 3.411624e-01 119.3624969
#>
#> $changedisc
#> LG PC
#> Macrophytes NA NA
#> Susp/dep feeding benthos -0.0008184394 -0.1882752
#> Carn/scav feeding benthos -0.1183588979 -23.8550507
#> Pelagic invertebrates NA NA
#> Planktivorous fish 0.0098975832 2.3051704
#> Migratory fish -0.0193985593 -4.3683959
#> Non-quota demersal fish -0.0332555585 -7.3715405
#> Quota-limited demersal fish -0.0857658131 -17.9205974
#> Birds 0.0239826045 5.6775180
#> Pinnipeds -0.0042272205 -0.9686317
#> Cetaceans 0.3157041768 106.8731735
#>
# }